STA 142A Statistical Learning I
Discussion 8: A Short Tutorial in pyGAM
TA: Tesi Xiao
Recap
Generalized Additive Models (GAMs) are smooth semi-parametric models of the form:
\[g(\mathbb{E}[Y|X]) = \beta_0 + f_1(X_1) + f_2(X_2) + \dots + f_d(X_d)\]where $X = [X_1, X_2, \dots, X_d]^\top, Y\in\mathbb{R}$, and $g()$ is the link function that relates the predictor variables to the conditioned expected value of $Y$.
The functions $f_i(\cdot)$ are built using penalized basis spline (B-spline), also known as P-spline, which can be interpreted as smoothing spline with specific basis functions. These functions $f_i$’s allow us to automatically model non-linear relationships without having to manually try out many different transformations on each variable.
In general, we can express all kinds of Generalized Additive Models in the form of
\[Y|X \sim \text{ExponentialFamily}(\mu),\quad \text{where }\mu = \mathbb{E}[Y|X]\]and
\[g(\mu) = \beta_0 + f_1(X_1) + f_2(X_2) + \dots + f_d(X_d)\]Remark. In general, each function $f_i$ in GAM can be a multi-variate function such as $f_i(X_p, X_q)$.
pyGAM
Create model instances
From the above formulation, we can see that a GAM consists of 3 components:
distributionfrom the exponential familylink function$g(\cdot)$function formwith an additive structure $\beta_0 + f_1(X_1) + f_2(X_2) + \dots + f_d(X_d)$
In pyGAM, we can use
class pygam.pygam.GAM(terms='auto', max_iter=100, tol=0.0001, distribution='normal', link='identity', callbacks=['deviance', 'diffs'], fit_intercept=True, verbose=False, **kwargs)
to create a GAM specified by
distribution:'normal','binomial','poisson','gamma','inv_gauss'link:'identity','logit','inverse','log','inverse-squared'terms: we can either choose ‘auto’ by default (without passing any argument toterms) os specify the functional form manually by passing an expression toterms. To be specific, we specify the functional form using terms:l()- linear terms;s()- spline terms;f()- factor terms;intercept.
Remark. Note that models include an intercept by default due to fit_intercept=True.
For example, if we want to fit a linear GAM (distribution='normal', link='identity') like
where $X_1, X_2$ are numeric variables, $X_3$ are binary-valued variables ($X_3=0,1$). Then we can build the model using GAM like:
from pygam import GAM, s, l, f
GAM(s(0) + l(1) + f(2))
GAM(callbacks=['deviance', 'diffs'], distribution='normal',
fit_intercept=True, link='identity', max_iter=100,
terms=s(0) + l(1) + f(2), tol=0.0001, verbose=False)
Remark.
distribution='normal', link='identity'andfit_intercept=Trueare already set by default. The index of the preditor variable starts at 0, i.e., $0 –> X_1; 1–>X_2; 2–>X_3$- we can also specify different arguments in terms
s(),l(), andf(); see API. For instance, we can specify the order of basis functions and the penalty parameterlamin the B-spline term.
pygam.terms.s(feature, n_splines=20, spline_order=3, lam=0.6, penalties='auto', constraints=None, dtype='numerical', basis='ps', by=None, edge_knots=None, verbose=False)
In pyGAM, for convenience, you can build custom models by specifying these 3 elements, or you can choose from common models:
LinearGAMidentity link and normal distributionLogisticGAMlogit link and binomial distributionPoissonGAMlog link and Poisson distributionGammaGAMlog link and gamma distributionInvGausslog link and inv_gauss distribution
Fit models
In pyGAM, you can either use .fit() or .gridsearch() to fit GAMs on the training set. The difference is that .gridsearch() performs a grid search over a space of all tunable parameters; see API. Thus, .gridsearch() is slow but may have better results.
from pygam import LinearGAM, s
from pygam.datasets import toy_interaction
X, y = toy_interaction(return_X_y=True)
gam = LinearGAM(s(0)).fit(X, y)
gam.summary()
LinearGAM
=============================================== ==========================================================
Distribution: NormalDist Effective DoF: 19.1074
Link Function: IdentityLink Log Likelihood: -123123.6702
Number of Samples: 50000 AIC: 246287.5553
AICc: 246287.5722
GCV: 4.1528
Scale: 4.15
Pseudo R-Squared: 0.0005
==========================================================================================================
Feature Function Lambda Rank EDoF P > x Sig. Code
================================= ==================== ============ ============ ============ ============
s(0) [0.6] 20 19.1 9.06e-02 .
intercept 1 0.0 8.54e-01
==========================================================================================================
Significance codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
WARNING: Fitting splines and a linear function to a feature introduces a model identifiability problem
which can cause p-values to appear significant when they are not.
WARNING: p-values calculated in this manner behave correctly for un-penalized models or models with
known smoothing parameters, but when smoothing parameters have been estimated, the p-values
are typically lower than they should be, meaning that the tests reject the null too readily.
The method .predict(X) predicts expected value of target given model and input $X$. The method score(X, y, weights=None) is to compute the explained deviance for a trained model for a given $X$ and $y$.
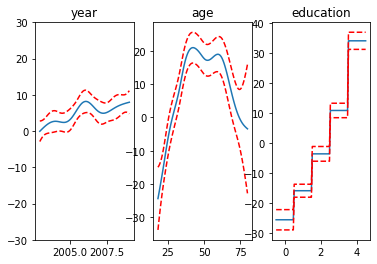
Visualize each function
from pygam import LinearGAM, s, f
from pygam.datasets import wage
import matplotlib.pyplot as plt
X, y = wage(return_X_y=True)
## model
gam = LinearGAM(s(0) + s(1) + f(2))
gam.gridsearch(X, y)
## plotting
plt.figure();
fig, axs = plt.subplots(1,3);
titles = ['year', 'age', 'education']
for i, ax in enumerate(axs):
XX = gam.generate_X_grid(term=i)
ax.plot(XX[:, i], gam.partial_dependence(term=i, X=XX))
ax.plot(XX[:, i], gam.partial_dependence(term=i, X=XX, width=.95)[1], c='r', ls='--')
if i == 0:
ax.set_ylim(-30,30)
ax.set_title(titles[i]);